U-net3+ network-based prediction of target and dangerous organ location in cervical cancer after-loading therapy
-
摘要:
目的 构建基于深度学习方法的宫颈癌后装治疗中高危靶区和危及器官位置预测方法。 方法 构建基于U-net3+的端到端自动分割框架,对两个中心213例已进行后装高剂量率治疗的宫颈癌患者进行勾画,并按照7:2:1的比例分为训练集、验证集和测试集。勾画的内容包括高危临床靶区、膀胱、直肠和小肠,分别用豪斯多夫距离及戴斯相似系数评估预测模型的准确性。 结果 膀胱自动勾画的戴斯相似系数为0.953,直肠、小肠分别为0.885、0.857,危及器官的平均值是0.898,豪斯多夫距离平均为5.4 mm;高危临床靶区戴斯相似系数是0.869,豪斯多夫距离为8.1 mm。 结论 基于U-net3+的宫颈癌后装治疗中靶区和危及器官位置预测模型具有较高的准确率,同时训练耗费时间少,有望在临床进行应用推广。 Abstract:Objective To conduct a model to predicate high-risk target areas and dangerous organ locations in after-loading therapy for cervical cancer based on deep learning. Methods An end-to-end automatic segmentation framework based on U-net3+ was constructed, and 213 cervical cancer patients who had undergone after-loading high-dose-rate therapy in the two centers were delineated. The patients were divided into training set, validation set and test set according to the ratio of 7:2:1. The high-risk clinical target, bladder, rectum, and small intestine were delineated, and the accuracy of the predictive model was assessed using the Hausdorff distance and the Dice similarity coefficient, respectively. Results The Dice similarity coefficient values of automatic bladder delineation were 0.953, rectum and small intestine were 0.885 and 0.857, respectively. The mean value of organ risk was 0.898, and the average Hausdorff distance value was 5.4 mm. The Dice similarity coefficient value of the high-risk clinical target was 0.869 and the Hausdorff distance value was 8.1 mm. Conclusion The U-net3+based model for predicting the location of target and dangerous organs in after-loading therapy for cervical cancer has a high accuracy rate, while the training is less time-consuming. It is expected to be promoted for clinical application. -
Key words:
- cervical cancer /
- after-loading therapy /
- location prediction /
- deep learning /
- U-net3+
-
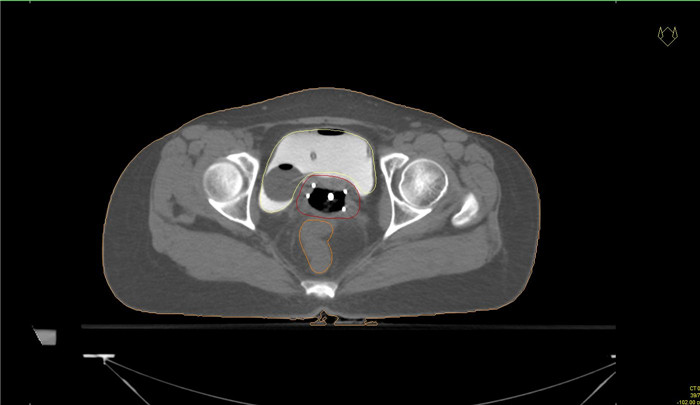
图 4 自动勾画结果
Figure 4. Automatic segmentation results.
A and E, B and F, C and G, D and H indicated the bladder, rectum, small intestine and HRCTV outline results respectively, while A and E indicated the bladder outline results of case 1 and case 2 respectively, B and F indicated the rectum outline results of case 1 and case 2 respectively, C and G indicated the small intestine outline results of case 1 and case 2 respectively, D and H indicated the HRCTV outline results of case 1 and case 2 respectively. The yellow line indicated the manually outlined contour line, and the red line indicated the automatically outlined result.
表 1 位置预测准确性结果
Table 1. Quantification of the accuracy of automatic segmentation
Structures DSC (Mean±SD) HD (mm, Mean±SD) Training time (h) Bladder 0.953±0.020 5.1±1.6 8 Rectum 0.885±0.030 5.4±1.5 11 Small intestine 0.857±0.040 5.7±2.1 12 HRCTV 0.869±0.030 8.1±2.8 15 DSC: Dice similarity coefficient; HD: Hausdorff distance; HRCTV: High-risk clinical target. -
[1] Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics[J]. 2021 CA A Cancer J Clin, 2021, 71(1): 7-33. [2] Matsuo K, Machida H, Mandelbaum RS, et al. Validation of the 2018 FIGO cervical cancer staging system[J]. Gynecol Oncol, 2019, 152(1): 87-93. doi: 10.1016/j.ygyno.2018.10.026 [3] Jauniaux E, Ayres-de-Campos D, Langhoff-Roos J, et al. FIGO classification for the clinical diagnosis of placenta accreta spectrum disorders[J]. Int J Gynaecol Obstet, 2019, 146(1): 20-4. doi: 10.1002/ijgo.12761 [4] 吴德华. 子宫颈癌放射治疗的规范化[J]. 中国实用妇科与产科杂志, 2021, 37(1): 54-9. doi: 10.19538/j.fk2021010114 [5] Yang AN, Lu N, Jiang HY, et al. Automatic delineation of organs at risk in non-small cell lung cancer radiotherapy based on deep learning networks[J]. 肿瘤学与转化医学: 英文版, 2022(2): 83-8. https://www.cnki.com.cn/Article/CJFDTOTAL-ZDLZ202303003.htm [6] Chino J, Annunziata CM, Beriwal S, et al. Radiation Therapy for Cervical Cancer: Executive Summary of an ASTRO Clinical Practice Guideline[J]. Practical Radiation Oncology, 2020, 10(4): 220-34. doi: 10.1016/j.prro.2020.04.002 [7] Kleppe A, Skrede OJ, De Raedt S, et al. Designing deep learning studies in cancer diagnostics[J]. Nat Rev Cancer, 2021, 21(3): 199-211. doi: 10.1038/s41568-020-00327-9 [8] Ramesh AN, Kambhampati C, Monson J, et al. Artificial intelligence in medicine[J]. Ann R Coll Surg Engl, 2004, 86(5): 334-8. doi: 10.1308/147870804290 [9] Patel PR, Lei Y, Wang T, et al. Learning-based prostate needle position prediction for HDR brachytherapy[J]. Int J Radiat Oncol, 2021, 111(3): e290-1. [10] Akufuna E, MwiingaKalusopa V, Chitundu K, et al. Adherence to Radiation Therapy among Cervical Cancer Patients at Cancer Diseases Hospital in Lusaka, Zambia[J]. 生物科学与医学: 英文, 2022 (5): 25-39. https://www.cnki.com.cn/Article/CJFDTOTAL-ZXXY202319004.htm [11] 王先良, 罗锐, 黎杰, 等. 一种宫颈癌近距离治疗剂量预测系统: 中国, CN114596934A[P]. 2022-06-07. [12] 向艺达, 周剑良, 白雪, 等. 基于全卷积网络U-Net宫颈癌近距离治疗三维剂量分布预测研究[J]. 中华放射肿瘤学杂志, 2022, 31(4): 359-64. [13] 陈祥. 宫颈癌三维后装治疗插植针路径优化及施源器改进的剂量学研究[D]. 衡阳: 南华大学, 2019. [14] Guckenberger M, Lievens Y, Bouma AB, et al. Characterisation and classification of oligometastatic disease: a European Society for Radiotherapy and Oncology and European Organisation for Research and Treatment of Cancer consensus recommendation[J]. Lancet Oncol, 2020, 21(1): e18-28. doi: 10.1016/S1470-2045(19)30718-1 [15] Yin XX, Sun L, Fu YH, et al. U- net- based medical image segmentation[J]. J Healthc Eng, 2022, 2022: 1-16. [16] Huang HM, Lin LF, Tong RF, et al. UNet 3: a full-scale connected UNet for medical image segmentation[C]. ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 2020: 1055-9. [17] Krithika alias AnbuDevi M, Suganthi K. Review of semantic segmentation of medical images using modified architectures of UNET[J]. Diagnostics, 2022, 12(12): 3064. doi: 10.3390/diagnostics12123064 [18] Kosarac A, Cep R, Trochta M, et al. Thermal behavior modeling based on BP neural network in keras framework for motorized machine tool spindles[J]. Materials (Basel), 2022, 15(21): 7782. doi: 10.3390/ma15217782 [19] Grossman CD, Bari BA, Cohen JY. Serotonin neurons modulate learning rate through uncertainty[J]. Curr Biol, 2022, 32(3): 586-99. [20] Rino Neto J, Silva FPLD, Chilvarquer I, et al. Hausdorff Distance evaluation of orthodontic accessories'streaking artifacts in 3D model superimposition[J]. Braz Oral Res, 2012, 26(5): 450-6. [21] Tanabe Y, Ishida T, Eto H, et al. Evaluation of the correlation between prostatic displacement and rectal deformation using the Dice similarity coefficient of the rectum[J]. Med Dosim, 2019, 44 (4): e39-43. [22] Renganathan V. Overview of artificial neural network models in the biomedical domain[J]. Bratislava Med J, 2019, 120(7): 536-40. [23] Chiu YC, Zheng SY, Wang LJ, et al. Predicting and characterizing a cancer dependency map of tumors with deep learning[J]. Sci Adv, 2021, 7(34): eabh1275. [24] Mayr D, Schmoeckel E, Höhn AK, et al. Aktuelle WHO-klassifikation des weiblichen genitale[J]. Pathologe, 2021, 42(3): 259-69. [25] Wang JZ, Lu JY, Qin G, et al. Technical Note: a deep learning-based autosegmentation of rectal tumors in MR images[J]. Med Phys, 2018, 45(6): 2560-4. [26] Bera K, Braman N, Gupta A, et al. Predicting cancer outcomes with radiomics and artificial intelligence in radiology[J]. Nat Rev Clin Oncol, 2022, 19(2): 132-46. -
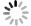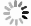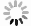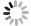






 下载:
下载: