Clinical prognosis of m6A RNA methylation-regulated genes in thyroid cancer
-
摘要:
目的探索m6A RNA甲基化调节基因对THCA的与后续影响。 方法分析癌症基因组图谱(TCGA)数据库中的RNA-seq数据及相关临床信息。通过wilcoxon检验分析肿瘤与正常样品之间m6A RNA甲基化调节基因的不同表达,使用lasso Cox回归分析构建m6A RNA甲基化调节基因与THCA的风险预后模型,使用中位风险评分将THCA患者分层为高风险组和低风险组,kaplan-Meier进行生存分析,使用ROC曲线用来评估预测模型的敏感性和特异性。 结果肿瘤与正常样品之间m6A RNA甲基化调节基因的不同表达,3个基因上调,14个基因下调。lasso Cox回归分析筛选出4个m6A RNA甲基化调节基因IGF2BP2、FTO、IGF2BP1、RBM15。高风险组与低风险组相比,OS明显较低(P=4.278e-02)。ROC曲线中AUC值为0.783,结果显示预测模型较有效。单因素和多因素Cox回归进行模型效能检验,单因素Cox回归分析中HR值为2.083(P < 0.001),多因素Cox回归分析中HR值为2.653(P < 0.001)。 结论m6A RNA甲基化调节基因IGF2BP2、FTO、IGF2BP1、RBM15可以构建预测模型来预测不同的临床亚群患者的OS。 -
关键词:
- m6A RNA甲基化 /
- 甲状腺癌 /
- 临床预后
Abstract:ObjectiveTo explore the correlation of m6A RNA methylation regulators and THCA. MethodsWe analyzed RNAseq data and related clinical information in the Cancer Genome Atlas (TCGA) database. The Wilcoxon test was used to analyze the different expressions of m6A RNA methylation-regulated genes between tumors and normal samples. A lasso Cox regression analysis was used to construct a risk prognosis model of m6A RNA methylation-regulated genes and THCA. The median risk score was used to divide THCA patients into high-risk group and low-risk group. Kaplan-Meier was performed for survival analysis. The ROC curve wa used to assess the sensitivity and feasibility of the prediction model. ResultsDifferent expression of m6A RNA methylation-regulated genes between tumors and normal samples showed 3 up-regulated genes and 14 down-regulated genes. Lasso Cox regression analysis screened four m6A RNA methylation regulating genes IGF2BP2, FTO, IGF2BP1 and RBM15. OS in the high-risk group was significantly lower than that in the low-risk group (P= 4.278e-02). The AUC value in the ROC curve was 0.783. The univariate and multivariate Cox regression were used to test the effectiveness of the model. The HR value in the univariate Cox regression analysis was 2.083, (P < 0.001). The HR value in the multivariate Cox regression analysis was 2.653 (P < 0.001). ConclusionThe m6A RNA methylation regulatory genes IGF2BP2, FTO, IGF2BP1 and RBM15 can construct predictive models to predict OS in patients with different clinical subgroups. -
Key words:
- m6A RNA methylation /
- thyroid cancer /
- clinical prognosis
-
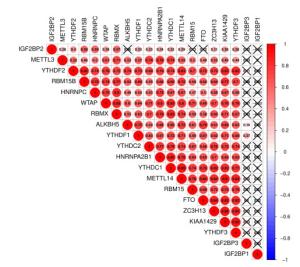
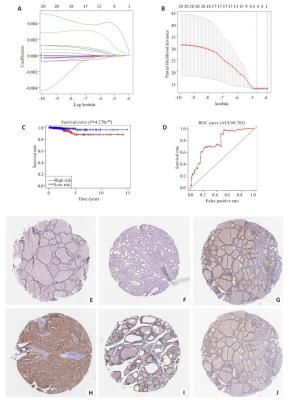
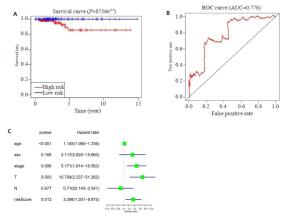
图 3 THCA中20个m6A RNA甲基化调节基因的风险特征
Figure 3. The risk signature of 20 m6A RNA methylation regulators in THCA. A, B: The coefficients calculated by Lasso Cox regression analysis; C: Kaplan-Meier overall survival curve for THCA patients between high risk and low risk group; D: ROC curve showed the predictive value of risk signature. E, G, I: The IHC expression of IGF2BP1、IGF2BP2、RBM15 in normal tissue; F, H, J: The IHC expression of IGF2BP1、IGF2BP2、RBM15 in tumor tissue.
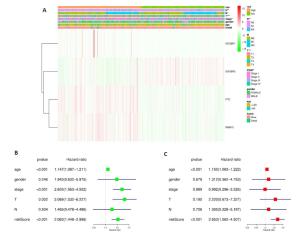
图 4 风险评分和临床特征对THCA患者预后的影响
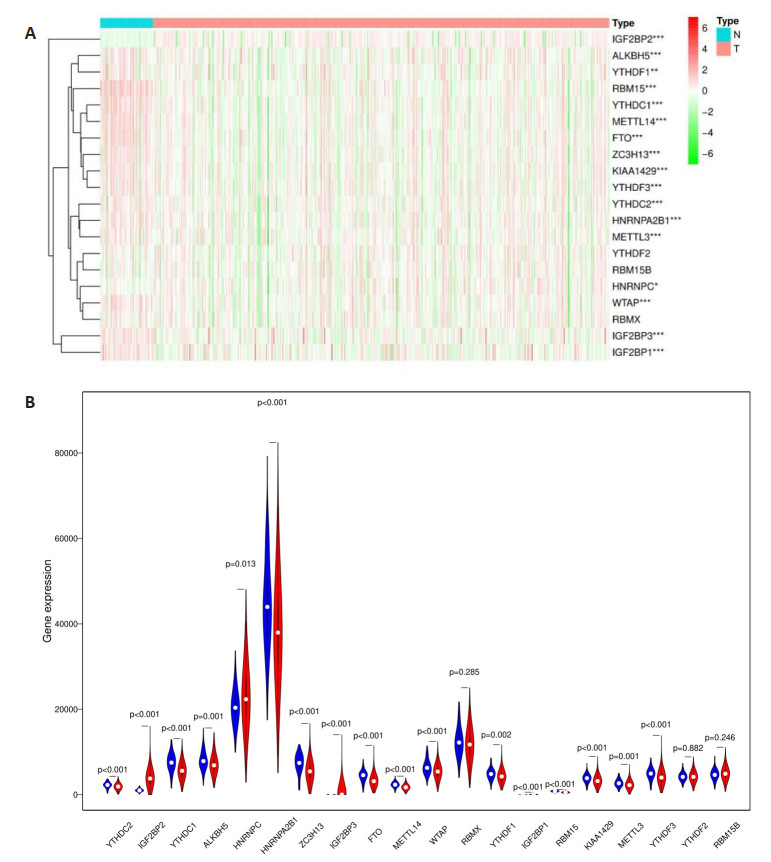
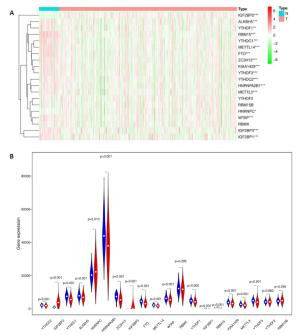
Figure 4. Effects of the risk signature and clinical information on the prognosis of CESC patients. A: The heat map showed the different expression of m6A RNA methylation regulators and the distribution of clinical information between high risk group and low risk group; Univariate Cox regression (B) and Multivariate Cox regression (C) analysis of the correlation between clinical information and overall survival.
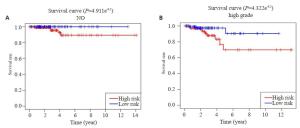
图 6 在验证集中验证预后模型
Figure 6. Validation of the prognostic signature in tested group. A: Kaplan-Meier overall survival curve for THCA patients between high risk and low risk group in tested group; B: ROC curve showed the predictive value of risk signature in tested group; C: Univariate Cox regression analysis the correlation between clinical information and overall survival in tested group. *P < 0.05.
表 1 风险分组临床特征(n, %)
Table 1. The clinical information in different risk group
Characterisitic High risk Low risk Age (years) ≤60 186 (75) 200 (80.32) >60 62 (25) 49 (19.68) Gender Male 64 (25.8) 70 (28.11) Female 184 (74.2) 179 (71.89) Stage Ⅰ 147 (59.27) 132 (53.01) Ⅱ 30 (12.10) 22 (8.83) Ⅲ 54 (21.77) 58 (23.29) Ⅳ 17 (6.86) 37 (14.87) -
[1] Fagin JA, Mitsiades N. Molecular pathology of thyroid cancer: diagnostic and clinical implications[J]. Best Pract Res Clin Endocrinol Metab, 2008, 22(6): 955-69. http://europepmc.org/articles/PMC2615540 [2] la Vecchia C, Malvezzi M, Bosetti C, et al. Thyroid cancer mortality and incidence: a global overview[J]. Int J Cancer, 2015, 136(9): 2187-95. https://pubmed.ncbi.nlm.nih.gov/25284703/ [3] Carling T, Udelsman R. Thyroid cancer[J]. Annu Rev Med, 2014, 65: 125-37. https://www.sciencedirect.com/science/article/pii/B9780123969675000165 [4] Cha YJ, Koo JS. Next-generation sequencing in thyroid cancer[J]. J Transl Med, 2016, 14(1): 322-8. https://pubmed.ncbi.nlm.nih.gov/27871285/ [5] Fan C, Wang W, Jin J, et al. RASSF10 is epigenetically inactivated and suppresses cell proliferation and induces cell apoptosis by activating the p53 signalling pathway in papillary thyroid carcinoma cancer[J]. Cell Physiol Biochem, 2017, 41(3): 1229-39. https://www.ncbi.nlm.nih.gov/pubmed/28268222 [6] Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019[J]. Cancer J Clin, 2019, 69(1): 7-34. http://www.ncbi.nlm.nih.gov/pubmed/30762872 [7] 杨雷, 孙婷婷, 袁延楠, 等. 1995-2010年北京城区甲状腺癌发病趋势及病理特征分析[J].中华预防医学杂志, 2013, 47(2): 109-12. http://d.wanfangdata.com.cn/Periodical/zhyfyx201302004 [8] Vigneri R, Malandrino P, Vigneri P. The changing epidemiology of thyroid cancer: why is incidence increasing[J]. Curr Opin Oncol, 2015, 27(1): 1-7. http://europepmc.org/abstract/med/25310641 [9] Jillard CL, Scheri RP, Sosa JA. What is the optimal treatment of papillary thyroid cancer[J].Adv Surg, 2015, 49(1): 79-93. http://www.sciencedirect.com/science/article/pii/S0065341115000081 [10] McLeod DS, Sawka AM, Cooper DS. Controversies in primary treatment of low-risk papillary thyroid cancer[J]. Lancet, 2013, 381 (9871): 1046-57. https://www.sciencedirect.com/science/article/pii/S0140673612622053 [11] Soares P, Celestino R, Melo M, et al. Prognostic biomarkers in thyroid cancer[J]. VirchowsArch, 2014, 464(3): 333-46. doi: 10.1007/s00428-013-1521-2 [12] Yapa S, Mulla O, Green V, et al. The role of chemokines in thyroid carcinoma[J]. Thyroid, 2017, 27(11): 1347-59. doi: 10.1089/thy.2016.0660 [13] Luo DY, Chen HB, Li XJ, et al. Activation of the ROCK1/MMP-9 pathway is associated with the invasion and poor prognosis in papillary thyroid carcinoma[J]. Int J Oncol, 2017, 51(4): 1209-18. https://www.spandidos-publications.com/ijo/51/4/1209 [14] Brennan K, Holsinger C, Dosiou C, et al. Development of prognostic signatures for intermediate-risk papillary thyroid cancer[J]. BMC Cancer, 2016, 16(1): 309-18. http://europepmc.org/articles/PMC5025616/ [15] Martínez J, Vargas-Salas S, Gamboa SU, et al. The combination of RET, BRAF and demographic data identifies subsets of patients with aggressive papillary thyroid cancer[J]. Horm Cancer, 2019, 10(23): 97-106. doi: 10.1007/s12672-019-0359-8 [16] Hua W, Zhao Y, Jin X, et al. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition[J]. Gynecol Oncol, 2018, 151 (2): 356-65. https://www.sciencedirect.com/science/article/pii/S0090825818312344 [17] Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control [J]. Nature, 2017, 552(7683): 126-31. http://europepmc.org/abstract/MED/29186125 [18] Lin X, Chai G, Wu Y, et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail[J]. Nat Commun, 2019, 10(1): 2065-73. http://www.nature.com/articles/s41467-019-09865-9 [19] Zhang C, Samanta D, Lu H, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6 A-demethylation of NANOG mRNA[J]. Proc Natl Acad Sci, 2016, 113(14): E2047-56. http://europepmc.org/abstract/MED/27001847 [20] Niu Y, Lin Z, Wan A, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3 [J]. Mol Cancer, 2019, 18(1): 46-53. https://pubmed.ncbi.nlm.nih.gov/30922314/ [21] Killock D. Genetics: the cancer genome atlas maps papillary thyroid cancer[J]. Nat Rev Clin Oncol, 2014, 11(12): 2150-8. https://www.ncbi.nlm.nih.gov/pubmed/25384944 [22] Yoo SK, Song YS, Lee EK, et al. Integrative analysis of genomic and transcriptomic characteristics associated with progression of aggressive thyroid cancer[J]. Nat Commun, 2019, 10(1): 1-12. https://www.ncbi.nlm.nih.gov/pubmed/?term=31235699 [23] Santarpia L, Calin GA, Adam L, et al. A miRNA signature associated with human metastatic medullary thyroid carcinoma[J]. Endocr Relat Cancer, 2013, 20(6): 809-23. https://moh-it.pure.elsevier.com/en/publications/a-mirna-signature-associated-with-human-metastatic-medullary-thyr [24] Han LO, Li XY, Cao MM, et al. Development and validation of an individualized diagnostic signature in thyroid cancer[J]. Cancer Med, 2018, 7(4): 1135-40. doi: 10.1002/cam4.1397 [25] Kim K, Jeon S, Kim T, et al. Immune gene signature delineates a subclass of papillary thyroid cancer with unfavorable clinical outcomes[J]. Cancers, 2018, 10(12): 494-9. https://www.ncbi.nlm.nih.gov/pubmed/30563160 [26] Teng HJ, Mao FB, Liang JL, et al. Transcriptomic signature associated with carcinogenesis and aggressiveness of papillary thyroid carcinoma[J]. Theranostics, 2018, 8(16): 4345-58. http://www.thno.org/v08p4345.htm [27] Wang ZY, Lv J, Zou X, et al. A three plasma microRNA signature for papillary thyroid carcinoma diagnosis in Chinese patients[J]. Gene, 2019, 693: 37-45. https://www.sciencedirect.com/science/article/pii/S0378111919300605 [28] Li QY, Wang P, Sun CH, et al. Integrative analysis of methylation and transcriptome identified epigenetically regulated lncRNAs with prognostic relevance for thyroid cancer[J]. Front Bioeng Biotechnol, 2020, 7: 439-45. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6962111/ [29] Yu WS, Zhao SJ, Wang YC, et al. Identification of cancer prognosisassociated functional modules using differential co-expression networks[J]. Oncotarget, 2017, 8(68): 112928-41. https://www.ncbi.nlm.nih.gov/pubmed/29348878 [30] Wang SW, Sun CX, Li JH, et al. Roles of RNA methylation by means of N6-methyladenosine (m6A) in human cancers[J]. Cancer Lett, 2017, 408: 112-20. https://www.sciencedirect.com/science/article/pii/S0304383517305177 [31] Wang KJ, Jiang L, Zhang Y, et al. Progression of thyroid carcinoma is promoted by the m6A methyltransferase METTL3 through regulating m6A methylation on TCF1[J]. Oncotargets Ther, 2020, 13: 1605-12. https://www.ncbi.nlm.nih.gov/pubmed/32158230 [32] Gao TY, Liu XX, He BS, et al. Long non-coding RNA 91H regulates IGF2 expression by interacting with IGF2BP2 and promotes tumorigenesis in colorectal cancer[J]. Artif Cells Nanomed Biotechnol, 2020, 48(1): 664-71. doi: 10.1080/21691401.2020.1727491 [33] Chen H, Kong Y, Yao Q, et al. Three hypomethylated genes were associated with poor overall survival in pancreatic cancer patients [J].Aging (Albany NY), 2019, 11(3): 885-97. https://www.ncbi.nlm.nih.gov/pubmed/30710069 [34] Liu H, Zeng Z, Afsharpad M, et al. Overexpression of IGF2BP3 as a potential oncogene in ovarian clear cell carcinoma[J]. Front Oncol, 2019, 9: 1570-8. https://www.ncbi.nlm.nih.gov/pubmed/32083017 [35] Zhang YY, Liu X, Liu LW, et al. Expression and prognostic significance of m6A-related genes in lung adenocarcinoma[J]. Med Sci Monit, 2020, 26: 644-52. https://pubmed.ncbi.nlm.nih.gov/32086933/ [36] Liu T, Li CY, Jin LP, et al. The prognostic value of m6A RNA methylation regulators in colon adenocarcinoma[J]. Med Sci Monit, 2019, 25: 9435-45. https://pubmed.ncbi.nlm.nih.gov/31823961 [37] Zhuang JF, Lin CL, Ye JX. M 6 A RNA methylation regulators contribute to malignant progression in rectal cancer[J]. J Cell Physiol, 2020, 235(9): 6300-6. doi: 10.1002/jcp.29626 [38] Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNAm6Amodification[J]. Cell Stem Cell, 2018, 22(2): 191-205. https://www.sciencedirect.com/science/article/pii/S193459091730468X [39] Xu F, Li CH, Wong CH, et al. Genome-wide screening and functional analysis identifies tumor suppressor long noncoding RNAs epigenetically silenced in hepatocellular carcinoma[J]. Cancer Res, 2019, 79(7): 1305-17. https://cancerres.aacrjournals.org/content/79/7/1305 [40] Müller S, Glaß M, Singh AK, et al. IGF2BP1 promotes SRFdependent transcription in cancer in a m6A- and miRNA-dependent manner[J]. NucleicAcids Res, 2019, 47(1): 375-90. https://academic.oup.com/nar/article/47/1/375/5146196 [41] Pan Y, Ma P, Liu Y, et al. Multiple functions of m6A RNA methylation in cancer[J]. J Hematol Oncol, 2018, 11(1): 48-56. doi: 10.1186/s13045-018-0590-8.pdf -





 下载:
下载: