Screening and identification of aptamers of liver cancer serum specific marker Dickkopf-1
-
摘要:
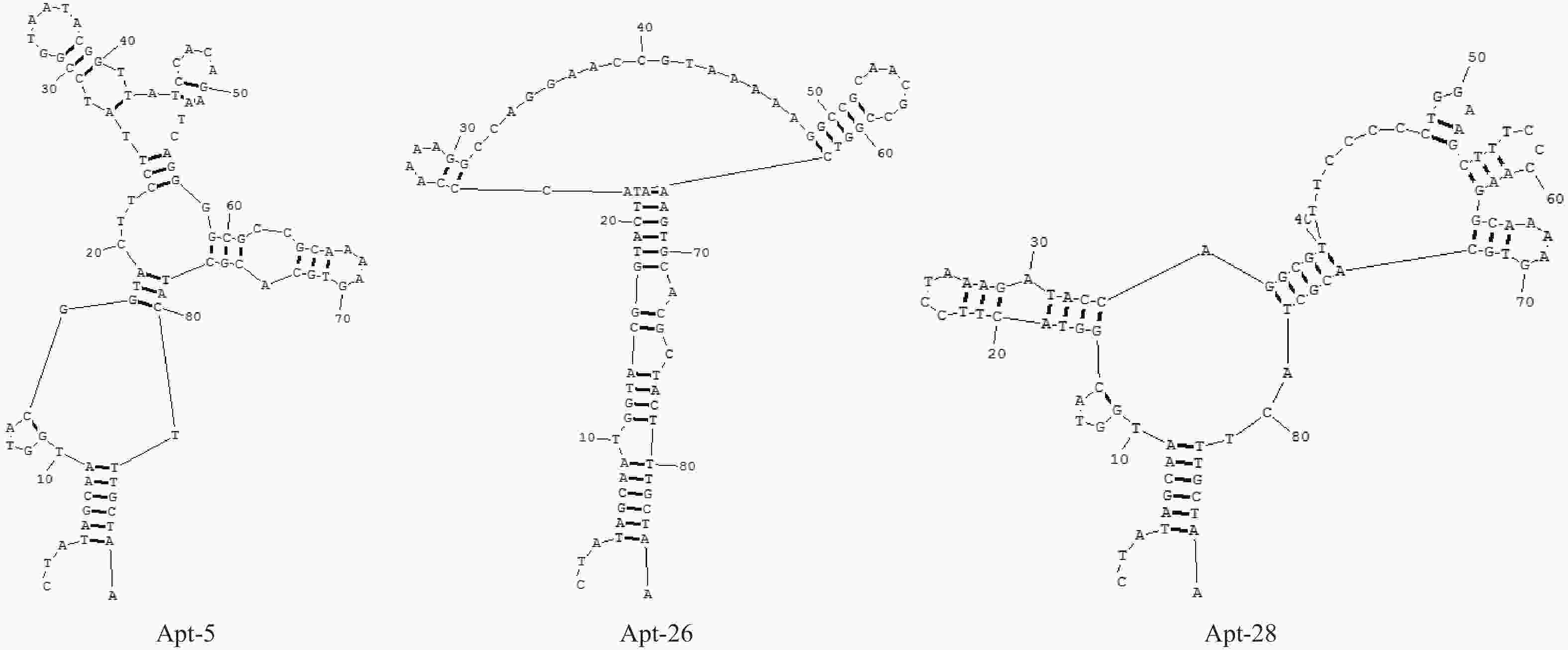
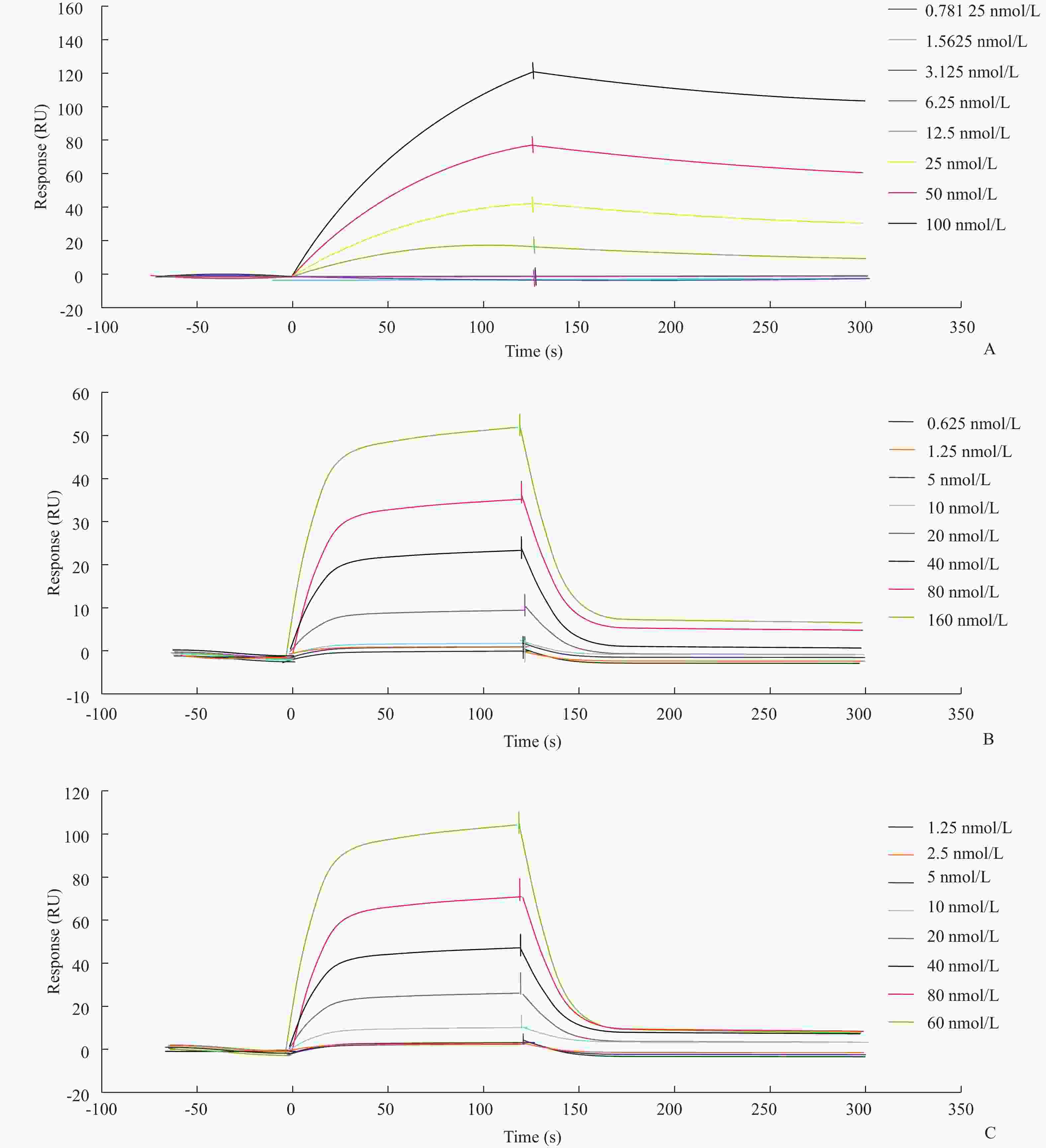
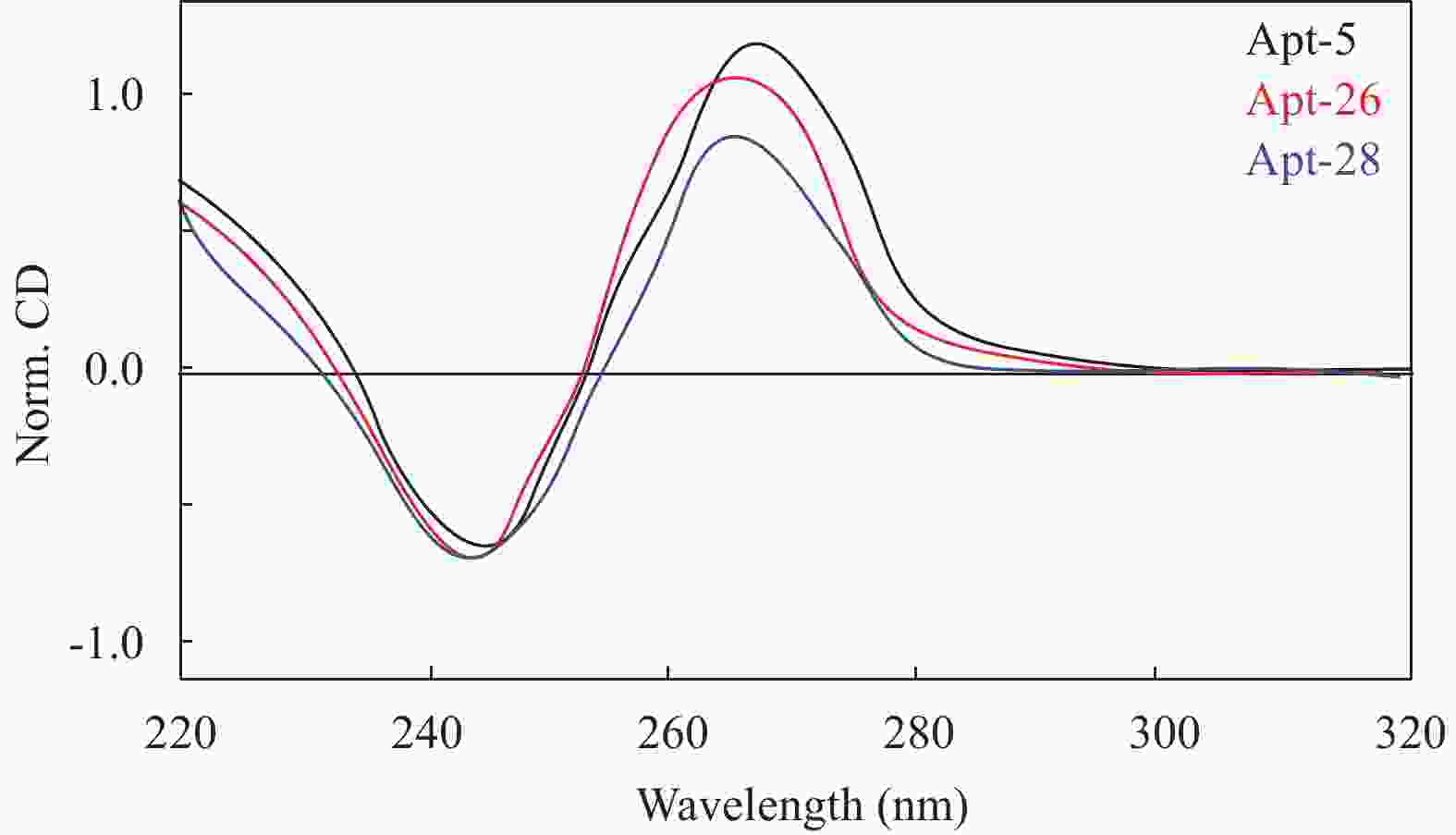
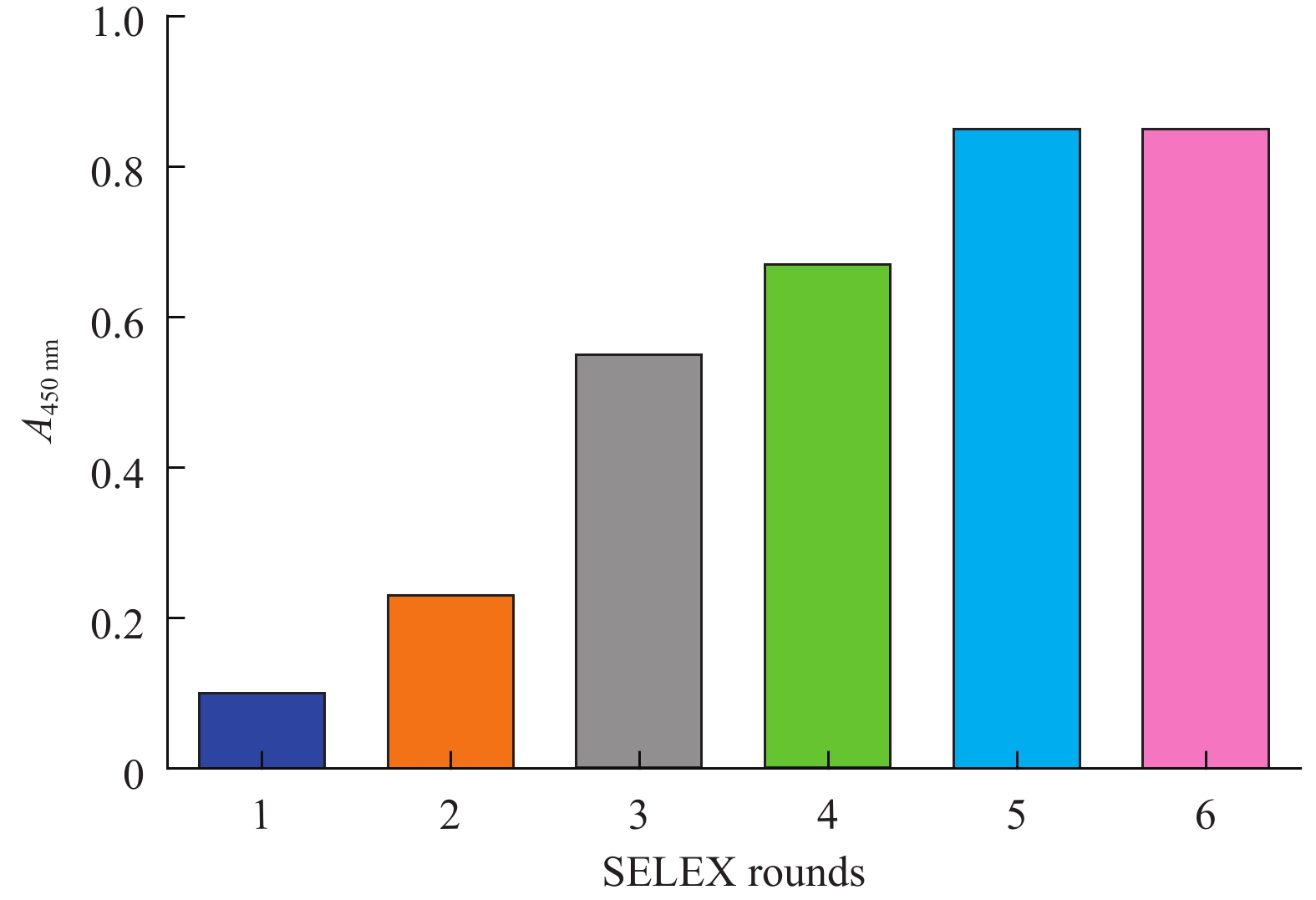
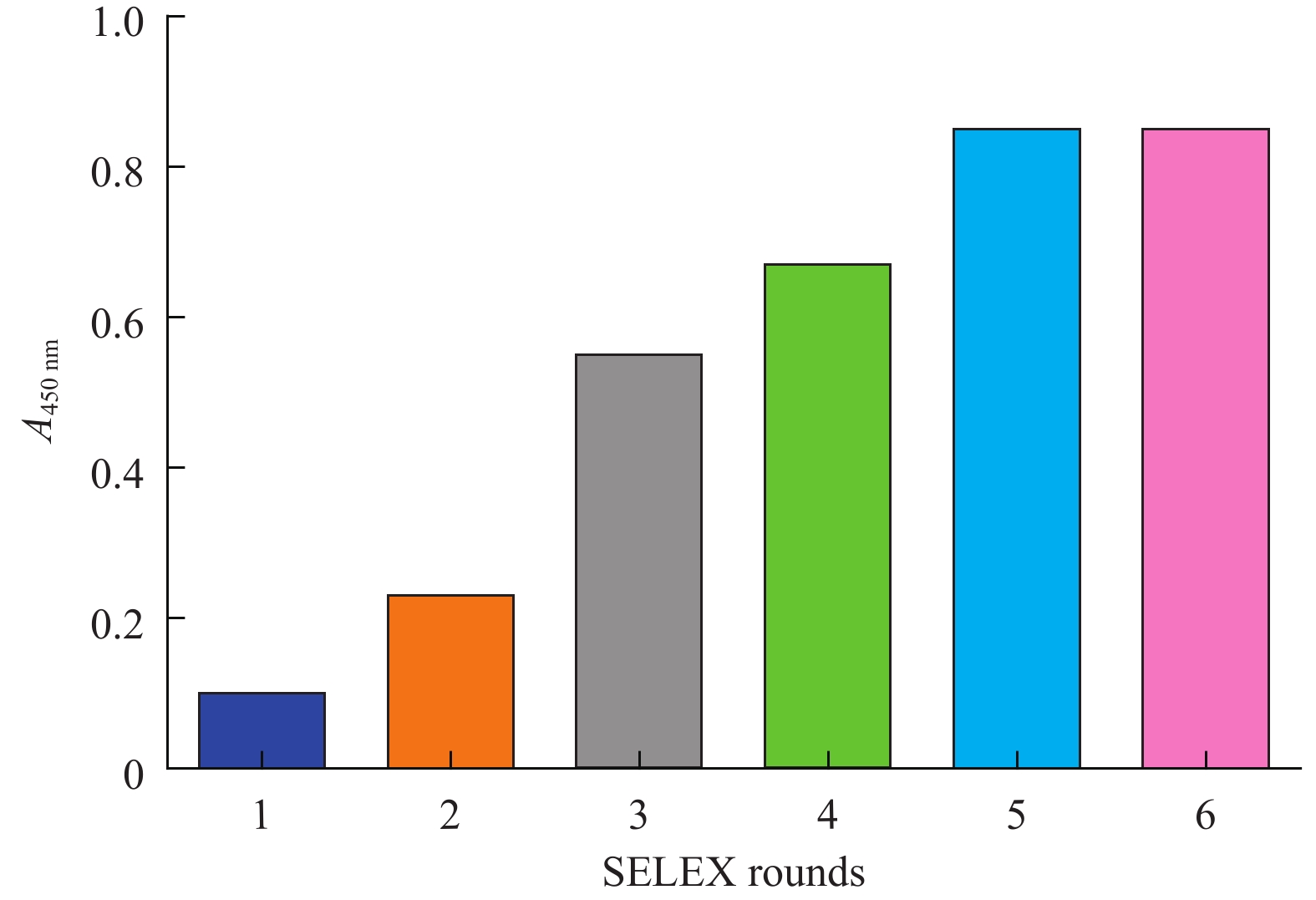
目的筛选和鉴定肝癌血清特异标志物Dickkopf-1核酸适体(Apt)。 方法采用配体指数级富集系统进化(SELEX)技术,以DKK1为靶蛋白,羧基化琼脂磁珠为筛选介质,从随机ssDNA文库中筛选出一组能够特异性与其结合的核酸适体。利用生物信息学方法对核酸适体进行序列分析和二级结构预测,表面等离子体共振仪分析测定核酸适体的亲和力。 结果经过6轮消减SELEX筛选,次级ssDNA文库与DKK1靶标蛋白的亲和力趋向稳定,将第 6 轮筛选产物经PCR扩增进行高通量测序。表面等离子体共振仪检测结果表明,筛选得到的DKK1核酸适体与DKK1的结合解离常数均在纳摩尔级水平,Apt-5的Kd值最小,亲和力最高,Apt-26和Apt-28核酸适体亲和力相对较弱。二级结构预测分析表明,茎环和茎凸环结构为主要的结构形式。圆二色光谱分析结果显示,3个候选核酸适体(Apt-5, Apt-26, Apt-28)能特异形成G-四链体结构识别DKK1靶标蛋白。 结论获得了与DKK1靶标蛋白特异性结合的核酸适体,为后续核酸适体的应用研究以及 DKK1蛋白功能的研究奠定了基础。 -
关键词:
- Dickkopf-1 /
- 配体指数级富集系统进化技术 /
- 核酸适体 /
- 亲和力
Abstract:ObjectiveTo screen and identify the serum specific marker Dickkopf-1 aptamer (Apt). MethodsThe systemic evolution of ligand by exponential enrichment (SELEX) was used to select DKK1 as the target protein and carboxylated agar magnetic beads as the screening medium. A random set of random ssDNA library was selected. A nucleic acid aptamer to which it binds. Sequence analysis and secondary structure prediction of nucleic acid aptamers were carried out by bioinformatics methods. The affinity of nucleic acid aptamers was determined by surface plasmon resonance (SPR) analysis. ResultsAfter 6 rounds of SELEX screening, the affinity of the secondary ssDNA library to the DKK1 target protein was stabilized. The 6th round of screening products were amplified by PCR for high-throughput sequencing. The results of SPR assay showed that the binding dissociation constants of DKK1 aptamers and DKK1 were in the nanomolar range. The Kd value of Apt-5 was the smallest and the affinity was the highest. The affinity of Apt-26 and Apt-28 aptamers was relative weak. The predictive analysis of secondary structure indicated that the stem loop and stem loop structure were the main structural forms. The results of circular dichroism spectroscopy showed that the three candidate aptamers (Apt-5, Apt-26, Apt-28) specifically formed the G-quadruplex structure to recognize the DKK1 target protein. Conclusion The aptamer that specifically binds to the DKK1 target protein is obtained. It laids a foundation for the application of subsequent aptamers and the study of DKK1 protein function. -
Key words:
- Dickkopf-1 /
- SELEX technique /
- nucleic acid aptamer /
- affinity
-
表 1 核酸适体测序结果
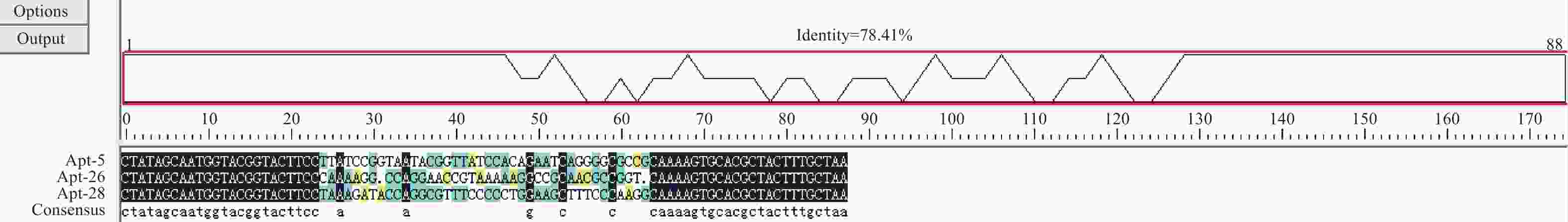
核酸适体 大小(bp) 序列(5′→3′) Kd(nmol·L) ΔG(kcal·mol−1) Apt-5 40 TTATCCGGTAATACGGTTATCCACAGAATCAGGGGCGCCG 12.26 −13.9 Apt-26 40 CAAAAGGCCAGGAACCGTAAAAAGGCCGCAACGCCGGTAA 24.85 −12.2 Apt-28 40 TAAAGATACCAGGCGTTTCCCCCTGGAAGCTTTCCCAAGG 21.18 −10.2 -
[1] Kim MC, Frank G. Liver resection for cancer: new developments in prediction, prevention and management of postresectional liver failure[J]. J Hepatol, 2016, 65(6): 1217-31. doi: 10.1016/j.jhep.2016.06.006 [2] Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008[J]. Int J Cancer, 2010, 127(12): 2893-917. doi: 10.1002/ijc.25516 [3] Taketa K. Alpha-fetoprotein: reevaluation in hepatology[J]. Hepatology, 1990, 12(6): 1420-32. doi: 10.1002/hep.1840120625 [4] Farinati F, Marino D, De Giorgio M, et al. Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither[J]. Am J Gastroenterol, 2006, 101(3): 524-32. doi: 10.1111/j.1572-0241.2006.00443.x [5] El-Serag HB, Marrero JA, Rudolph L, et al. Diagnosis and treatment of hepatocellular carcinoma[J]. Gastroenterology, 2008, 134(12): 1752-63. [6] Glinka A, Wu W, Delius H, et al. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction[J]. Nature, 1998, 391(6665): 357-62. doi: 10.1038/34848 [7] Yu B, Yang X, Xu Y, et al. Elevated expression of DKK1 is associated with cytoplasmic/nuclear beta-catenin accumulation and poor prognosis in hepatocellular carcinomas[J]. J Hepatol, 2009, 50(5): 948-57. doi: 10.1016/j.jhep.2008.11.020 [8] Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase[J]. Science, 1990, 249(4968): 505-10. doi: 10.1126/science.2200121 [9] Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands[J]. Nature, 1990, 346(6287): 818-22. doi: 10.1038/346818a0 [10] Shim WB, Kim MJ, Mun H, et al. An aptamer-based dipstick assay for the rapid and simple detection of aflatoxin B1[J]. Biosens Bioelectron, 2014, 62(5): 288-94. [11] Zheng J, Li J, Jiang Y, et al. Design of aptamer-based sensing platform using triple-helix molecular switch[J]. Anal Chem, 2011, 83(17): 6586-92. doi: 10.1021/ac201314y [12] Wang F, Banerjee D, Liu YS, et al. Upconversion nanoparticles in biological labeling, imaging and therapy[J]. Analyst, 2010, 135(8): 1839-54. doi: 10.1039/c0an00144a [13] Piyasena ME, Real LJ, Diamond RA, et al. Magnetic microsphere-based methods to study the interaction of teicoplanin with peptides and bacteria[J]. Anal Bioanal Chem, 2008, 392(5): 877-86. doi: 10.1007/s00216-008-2327-y [14] Karpik AE, Crulhas BP, Rodrigues CB, et al. Aptamer-based Biosensor developed to Monitor MUC1 released by prostate cancer cells[J]. Electroanalysis, 2017, 29(10): 2246-53. doi: 10.1002/elan.201700318 [15] Fang BY, Wang CY, Li C, et al. Amplified using DNase I and aptamer/graphene oxide for sensing prostate specific antigen in human serum[J]. Sens Actuators B Chem, 2017, 244(16): 928-33. [16] Cao S, Chen SJ. Physics-based de novo prediction of RNA 3D structures[J]. J Phys Chem B, 2011, 115(14): 4216-26. doi: 10.1021/jp112059y [17] Willner I, Zayats M. Electronic aptamer-based sensors[J]. Angew Chem Int Ed Engl, 2007, 46(34): 6408-18. doi: 10.1002/anie.200604524 [18] Bailor MH, Sun X, Al-Hashimi HM. Topology links RNA secondary structure with global conformation, dynamics, and adaptation[J]. Science, 2010, 327(5962): 202-6. doi: 10.1126/science.1181085 [19] Ahn JY, Lee S, Jo M, et al. Sol-gel derived nanoporous compositions for entrapping small molecules and their outlook toward aptamer screening[J]. Anal Chem, 2012, 84(6): 2647-53. doi: 10.1021/ac202559w [20] Ireson CR, Kelland LR. Discovery and development of anticancer aptamers[J]. Mol Cancer Ther, 2006, 5(12): 2957-62. doi: 10.1158/1535-7163.MCT-06-0172 [21] Sedighian H, Halabian R, Amani J, et al. Staggered target SELEX, a novel approach to isolate non-cross-reactive aptamer for detection of SEA by apta-qPCR[J]. J Biotechnol, 2018, 286(9): 45-55. [22] Zheng YE, Zhao YW, Di Y, et al. DNA aptamers from whole-serum SELEX as new diagnostic agents against gastric cancer[J]. RSC Adv, 2019, 9(2): 950-7. doi: 10.1039/C8RA08642G -





 下载:
下载: